Slopegraphs and R - a pleasant diversion
I try to at least scan the R-bloggers feed everyday. Not every article is of interest to me, but I often have one of two different reactions to at least one article. Sometimes it is an “ah ha” moment because the article is right on point for a problem I have now or have had in the past and the article provides a (better) solution. Other times my reaction is more of an “oh yeah”, because it is something I have been meaning to investigate, or something I once knew, but the article brings a different perspective to it.
The second case happened to me this week. I’ve been aware of slopegraphs and bumpcharts for quite some time, and I certainly am aware of Tufte’s work. As an amateur military historian I’ve always loved, for example, this poster depicting Napoleon’s Russian Campaign. So when I saw the article from Murtaza Haider titled “Edward Tufte’s Slopegraphs and political fortunes in Ontario” I just had to take a peek and revisit the topic.
The article does a good job of looking at slopegraphs in both R (via
plotrix) and Stata, even providing the code to do the work. My challenge was
that even though I’m adequate at plotting in base R, I much prefer using
ggplot2 wherever and whenever possible. My memory was that I had seen another
article on the related topic of a bumpchart on R-bloggers in the not too
distant past. A little sleuthing turned up this earlier
article from Dominik
Koch who wrote some code to compare
national performance at the Winter Olympics, “Bump Chart - Track performance
over time”.
Finally, I wound up at this Github
repository for a project called “Edward
Tufte-Inspired Slopegraphs” from Thomas J. Leeper
who has been building code to make slopegraphs using both base plotting
functions and ggplot2.
My post today will draw a little bit from all their work and hopefully provide
some useful samples for others to draw on if they share some of my quirks about
data layout and a preference for ggplot2 versus base plot. I’m going to
focus almost exclusively on slopegraphs, although much of the work could be
extended to bumpcharts as well.
Setup and library loading
We’re going to make occasional use of dplyr to manipulate the data, extensive
use of ggplot2 to do the plotting and ggrepel to solve one specific labeling
problem. We’ll load them and I am suppressing the message from dplyr about
namespace overrides.
require(dplyr)
require(ggplot2)
require(ggrepel)
require(kableExtra)Politics in Ontario
The original
post
is about plotting the data from some polling results in Ontario. For the
reader’s convenience I’ve made the data available via a structure command. We
have data about two different polling dates, for 5 political parties, and the
measured variable is percent of people supporting expressed as x.x (i.e. already
multiplied by 100).
data <- structure(list( Date = structure(c(1L, 1L, 1L, 1L, 1L, 2L, 2L, 2L, 2L, 2L),
.Label = c("11-May-18", "18-May-18"),
class = "factor"),
Party = structure(c(5L, 3L, 2L, 1L, 4L, 5L, 3L, 2L, 1L, 4L),
.Label = c("Green", "Liberal", "NDP", "Others", "PC"),
class = "factor"),
Pct = c(42.3, 28.4, 22.1, 5.4, 1.8, 41.9, 29.3, 22.3, 5, 1.4)),
class = "data.frame",
row.names = c(NA, -10L))
str(data)## 'data.frame': 10 obs. of 3 variables:
## $ Date : Factor w/ 2 levels "11-May-18","18-May-18": 1 1 1 1 1 2 2 2 2 2
## $ Party: Factor w/ 5 levels "Green","Liberal",..: 5 3 2 1 4 5 3 2 1 4
## $ Pct : num 42.3 28.4 22.1 5.4 1.8 41.9 29.3 22.3 5 1.4head(data)## Date Party Pct
## 1 11-May-18 PC 42.3
## 2 11-May-18 NDP 28.4
## 3 11-May-18 Liberal 22.1
## 4 11-May-18 Green 5.4
## 5 11-May-18 Others 1.8
## 6 18-May-18 PC 41.9Let’s just take the data as we have it and feed it to ggplot in a nice simple
fashion and see what we get with very little effort.
ggplot(data = data, aes(x = Date, y = Pct, group = Party)) +
geom_line(aes(color = Party, alpha = 1), size = 2) +
geom_point(aes(color = Party, alpha = 1), size = 4) +
# Labelling as desired
labs(
title = "Voter's stated preferences for June 7 elections in Ontario",
subtitle = "(Mainstreet Research)",
caption = "https://www.mainstreetresearch.ca/gap-between-ndp-and-pcs-narrows-while-liberals-hold-steady/"
)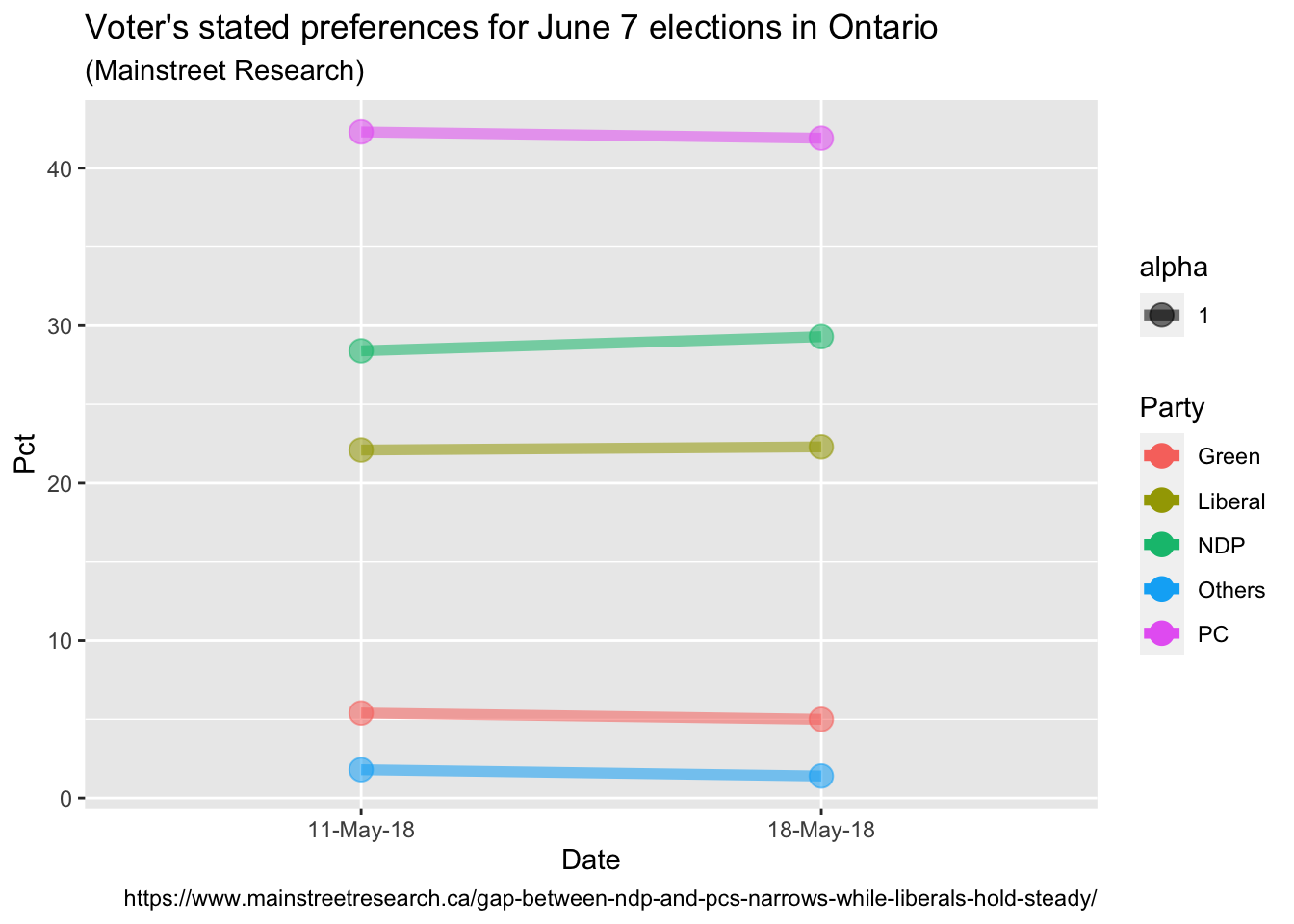
The nice thing about ggplot is once you get used to the syntax it becomes very
“readable”. We’ve identified our dataset, the x & y variables and our grouping
variable. Lines too big? An adjustment to size = 2 does it. Don’t like colors?
Pull the color = Party clause.
So we’re already pretty close to what we need. Things are scaled properly and
the basic labeling of titles etc. is accomplished. Our biggest “problem” is that
ggplot has been a little too helpful and adding some things we’d like to
remove to give it a more “Tuftesque” look. So what we’ll do in the next few
steps is add lines of code – but they are mainly designed to remove unwanted
elements. This is in contrast to a base plot where we have to write the code to
add elements.
So lets:
- Move the x axis labels to the top with
scale_x_discrete(position = "top") - Change to a nice clean black and white theme
theme_bw() - Not display any legend(s)
theme(legend.position = "none") - Remove the default border from our plot
theme(panel.border = element_blank())
ggplot(data = data, aes(x = Date, y = Pct, group = Party)) +
geom_line(aes(color = Party, alpha = 1), size = 2) +
geom_point(aes(color = Party, alpha = 1), size = 4) +
# move the x axis labels up top
scale_x_discrete(position = "top") +
theme_bw() +
# Format tweaks
# Remove the legend
theme(legend.position = "none") +
# Remove the panel border
theme(panel.border = element_blank()) +
# Labelling as desired
labs(
title = "Voter's stated preferences for June 7 elections in Ontario",
subtitle = "(Mainstreet Research)",
caption = "https://www.mainstreetresearch.ca/gap-between-ndp-and-pcs-narrows-while-liberals-hold-steady/"
)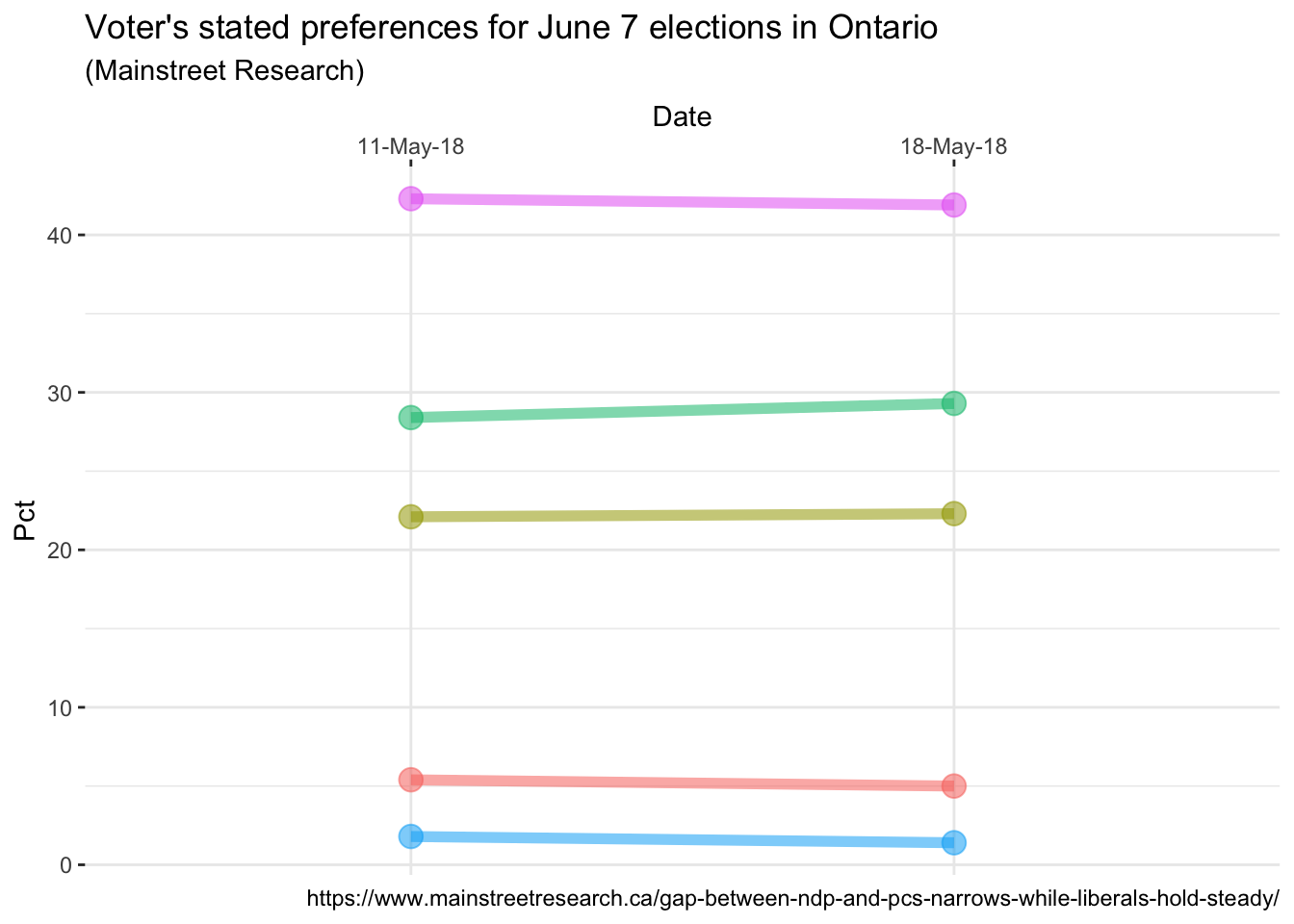
Nice progress! Continuing to remove things that can be considered “clutter” we
add some additional lines that all end in element_blank() and are invoked to
remove default plot items such as the plot grid, the y axis text, etc..
ggplot(data = data, aes(x = Date, y = Pct, group = Party)) +
geom_line(aes(color = Party, alpha = 1), size = 2) +
geom_point(aes(color = Party, alpha = 1), size = 4) +
# move the x axis labels up top
scale_x_discrete(position = "top") +
theme_bw() +
# Format tweaks
# Remove the legend
theme(legend.position = "none") +
# Remove the panel border
theme(panel.border = element_blank()) +
# Remove just about everything from the y axis
theme(axis.title.y = element_blank()) +
theme(axis.text.y = element_blank()) +
theme(panel.grid.major.y = element_blank()) +
theme(panel.grid.minor.y = element_blank()) +
# Remove a few things from the x axis and increase font size
theme(axis.title.x = element_blank()) +
theme(panel.grid.major.x = element_blank()) +
theme(axis.text.x.top = element_text(size=12)) +
# Remove x & y tick marks
theme(axis.ticks = element_blank()) +
# Labelling as desired
labs(
title = "Voter's stated preferences for June 7 elections in Ontario",
subtitle = "(Mainstreet Research)",
caption = "https://www.mainstreetresearch.ca/gap-between-ndp-and-pcs-narrows-while-liberals-hold-steady/"
)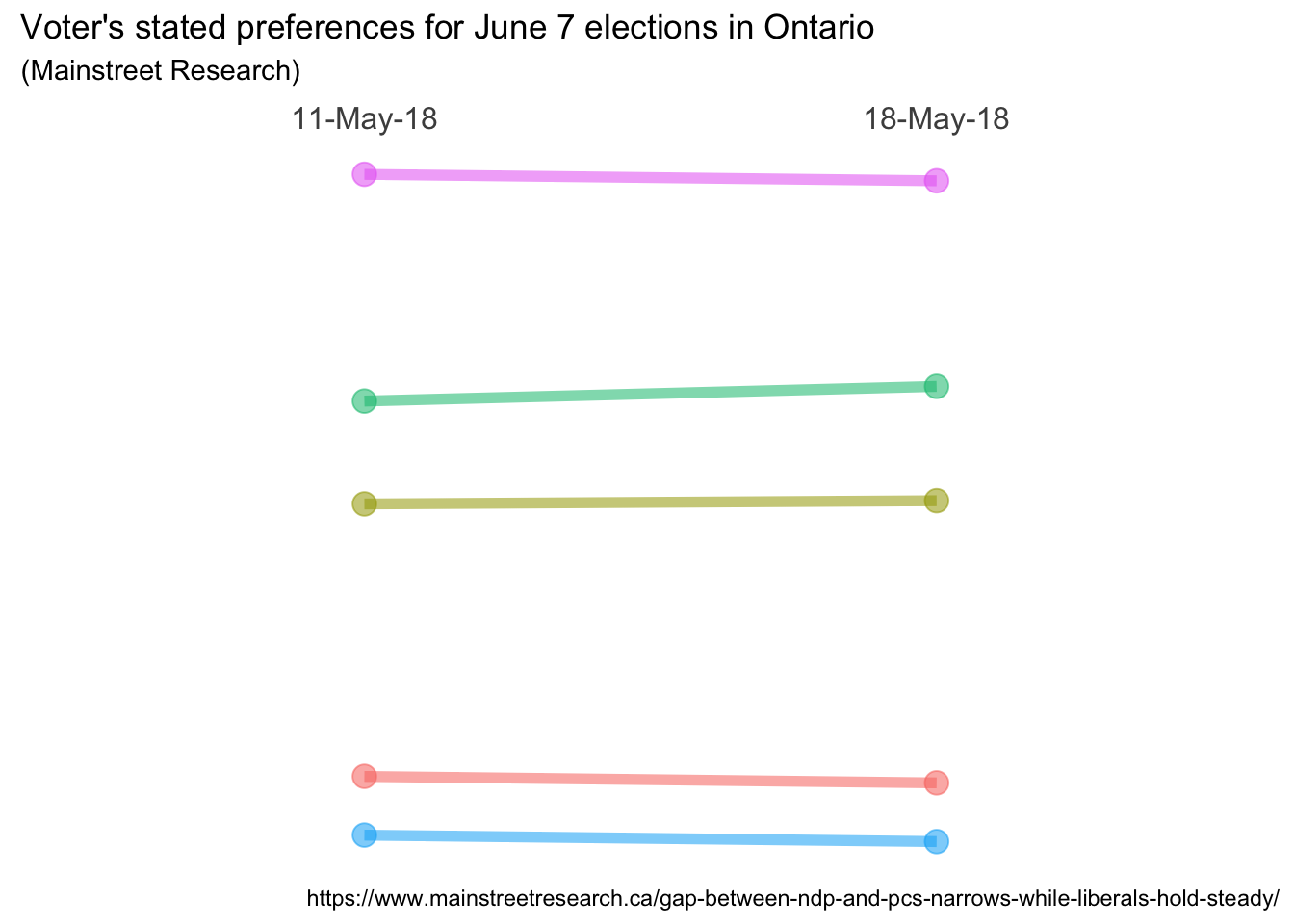
Very nice! We’re almost there! The “almost” is because now that we have removed
both the legend and all scales and tick marks we no longer know who is who, and
what the numbers are! Plus, I’m a little unhappy with the way the titles are
formatted, so we’ll play with that. Later, I’ll get fancy but for now let’s just
add some simple text labels on the left and right to show the party name and
their percentage. The code geom_text(aes(label = Party)) will place the party
name right on top of the points that anchor either end of the line. If we make
that geom_text(aes(label = paste0(Party, " - ", Pct, "%"))) then we’ll get
labels that have both the party and the percent all neatly formatted, but still
right on top of the points that anchor the ends of the line. hjust controls
horizontal justification so if we change it to geom_text(aes(label = paste0(Party, " - ", Pct, "%")), hjust = 1.35) both sets of labels will slide
to the left which is exactly what we want for the May 11 labels but not the May
18 labels. If we feed hjust a negative number they’ll go the other way. So
what we’ll do is filter the data using the filter function from dplyr and
place the left hand labels differently than the right hand labels. While we’re
at it we’ll make it bold face font and a little larger…
ggplot(data = data, aes(x = Date, y = Pct, group = Party)) +
geom_line(aes(color = Party, alpha = 1), size = 2) +
geom_point(aes(color = Party, alpha = 1), size = 4) +
geom_text(data = data %>% filter(Date == "11-May-18"),
aes(label = paste0(Party, " - ", Pct, "%")) ,
hjust = 1.35,
fontface = "bold",
size = 4) +
geom_text(data = data %>% filter(Date == "18-May-18"),
aes(label = paste0(Party, " - ", Pct, "%")) ,
hjust = -.35,
fontface = "bold",
size = 4) +
# move the x axis labels up top
scale_x_discrete(position = "top") +
theme_bw() +
# Format tweaks
# Remove the legend
theme(legend.position = "none") +
# Remove the panel border
theme(panel.border = element_blank()) +
# Remove just about everything from the y axis
theme(axis.title.y = element_blank()) +
theme(axis.text.y = element_blank()) +
theme(panel.grid.major.y = element_blank()) +
theme(panel.grid.minor.y = element_blank()) +
# Remove a few things from the x axis and increase font size
theme(axis.title.x = element_blank()) +
theme(panel.grid.major.x = element_blank()) +
theme(axis.text.x.top = element_text(size=12)) +
# Remove x & y tick marks
theme(axis.ticks = element_blank()) +
# Format title & subtitle
theme(plot.title = element_text(size=14, face = "bold", hjust = 0.5)) +
theme(plot.subtitle = element_text(hjust = 0.5)) +
# Labelling as desired
labs(
title = "Voter's stated preferences for June 7 elections in Ontario",
subtitle = "(Mainstreet Research)",
caption = "https://www.mainstreetresearch.ca/gap-between-ndp-and-pcs-narrows-while-liberals-hold-steady/"
)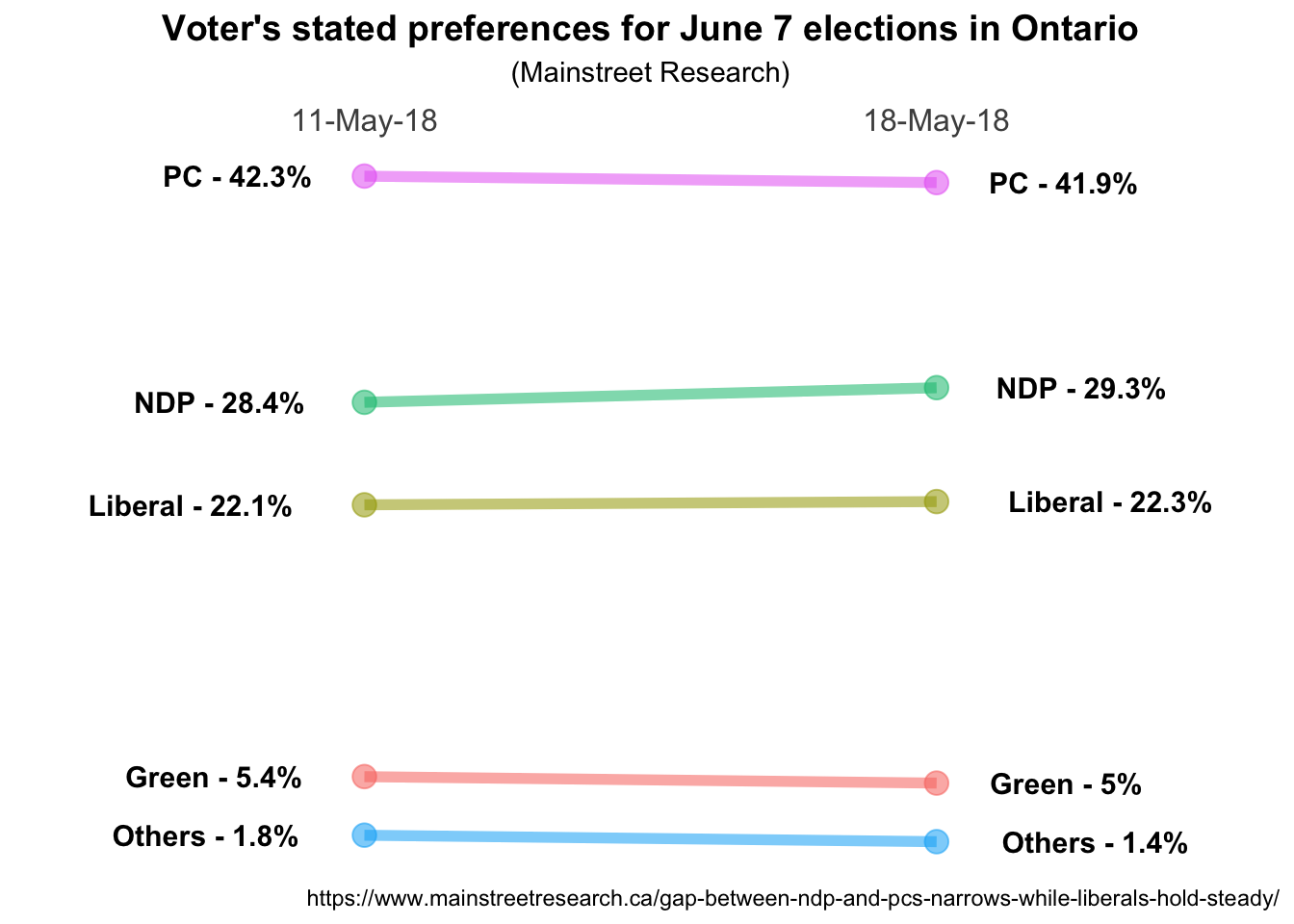
Eureka! Not perfect yet but definitely looking good.
Adding complexity
I’m feeling pretty good about the solution so far but there are three things I’d like to make better.
- How well will this solution work when we have more than two time periods? Need to make sure it generalizes to a more complex case.
- As Murtaza Haider notes in his post we’ll have issues if the data points are identical or very close together. Our very neat little labels will overlap each other. In his post I believe he mentions that he manually moved them in some cases. Let’s try and fix that.
- Oh my, that’s a lot of code to keep cutting and pasting, can we simplify?
To test #1 and #2 I have “invented”" a new dataset called moredata. It is
fictional it’s labelled May 25th but today is actually May 24th. But I created
it to add a third polling date and to make sure that we had a chance to test
what happens when we have two identical datapoints on the same day. Notice that
on May 25th the polling numbers for the Liberals and the NDP are identical at
26.8%.
moredata <- structure(list(Date = structure(c(1L, 1L, 1L, 1L, 1L, 2L, 2L, 2L, 2L, 2L, 3L, 3L, 3L, 3L, 3L),
.Label = c("11-May-18", "18-May-18", "25-May-18"),
class = "factor"),
Party = structure(c(5L, 3L, 2L, 1L, 4L, 5L, 3L, 2L, 1L, 4L, 5L, 3L, 2L, 1L, 4L),
.Label = c("Green", "Liberal", "NDP", "Others", "PC"),
class = "factor"),
Pct = c(42.3, 28.4, 22.1, 5.4, 1.8, 41.9, 29.3, 22.3, 5, 1.4, 41.9, 26.8, 26.8, 5, 1.4)),
class = "data.frame",
row.names = c(NA, -15L))
tail(moredata)## Date Party Pct
## 10 18-May-18 Others 1.4
## 11 25-May-18 PC 41.9
## 12 25-May-18 NDP 26.8
## 13 25-May-18 Liberal 26.8
## 14 25-May-18 Green 5.0
## 15 25-May-18 Others 1.4You’ll notice at the beginning of this post I loaded the ggrepel library.
ggrepel works with ggplot2 to repel things that overlap, in this case our
geom_text labels. The invocation is geom_text_repel and it is very similar
to geom_text but allows us to deconflict the overlaps. We’ll use hjust = "left" and hjust = "right" to control justifying the labels. We’ll use a
fixed nudge left and right nudge_x = -.45 and nudge_x = .5 to move the
labels left and right off the plotted data points and we will explicitly tell
geom_text_repel to only move the labels vertically to avoid overlap with
direction = "y". Everything else remains the same.
ggplot(data = moredata, aes(x = Date, y = Pct, group = Party)) +
geom_line(aes(color = Party, alpha = 1), size = 2) +
geom_point(aes(color = Party, alpha = 1), size = 4) +
geom_text_repel(data = moredata %>% filter(Date == "11-May-18"),
aes(label = paste0(Party, " - ", Pct, "%")) ,
hjust = "left",
fontface = "bold",
size = 4,
nudge_x = -.45,
direction = "y") +
geom_text_repel(data = moredata %>% filter(Date == "25-May-18"),
aes(label = paste0(Party, " - ", Pct, "%")) ,
hjust = "right",
fontface = "bold",
size = 4,
nudge_x = .5,
direction = "y") +
# move the x axis labels up top
scale_x_discrete(position = "top") +
theme_bw() +
# Format tweaks
# Remove the legend
theme(legend.position = "none") +
# Remove the panel border
theme(panel.border = element_blank()) +
# Remove just about everything from the y axis
theme(axis.title.y = element_blank()) +
theme(axis.text.y = element_blank()) +
theme(panel.grid.major.y = element_blank()) +
theme(panel.grid.minor.y = element_blank()) +
# Remove a few things from the x axis and increase font size
theme(axis.title.x = element_blank()) +
theme(panel.grid.major.x = element_blank()) +
theme(axis.text.x.top = element_text(size=12)) +
# Remove x & y tick marks
theme(axis.ticks = element_blank()) +
# Format title & subtitle
theme(plot.title = element_text(size=14, face = "bold", hjust = 0.5)) +
theme(plot.subtitle = element_text(hjust = 0.5)) +
# Labelling as desired
labs(
title = "Bogus Data",
subtitle = "(Chuck Powell)",
caption = "https://www.mainstreetresearch.ca/gap-between-ndp-and-pcs-narrows-while-liberals-hold-steady/"
)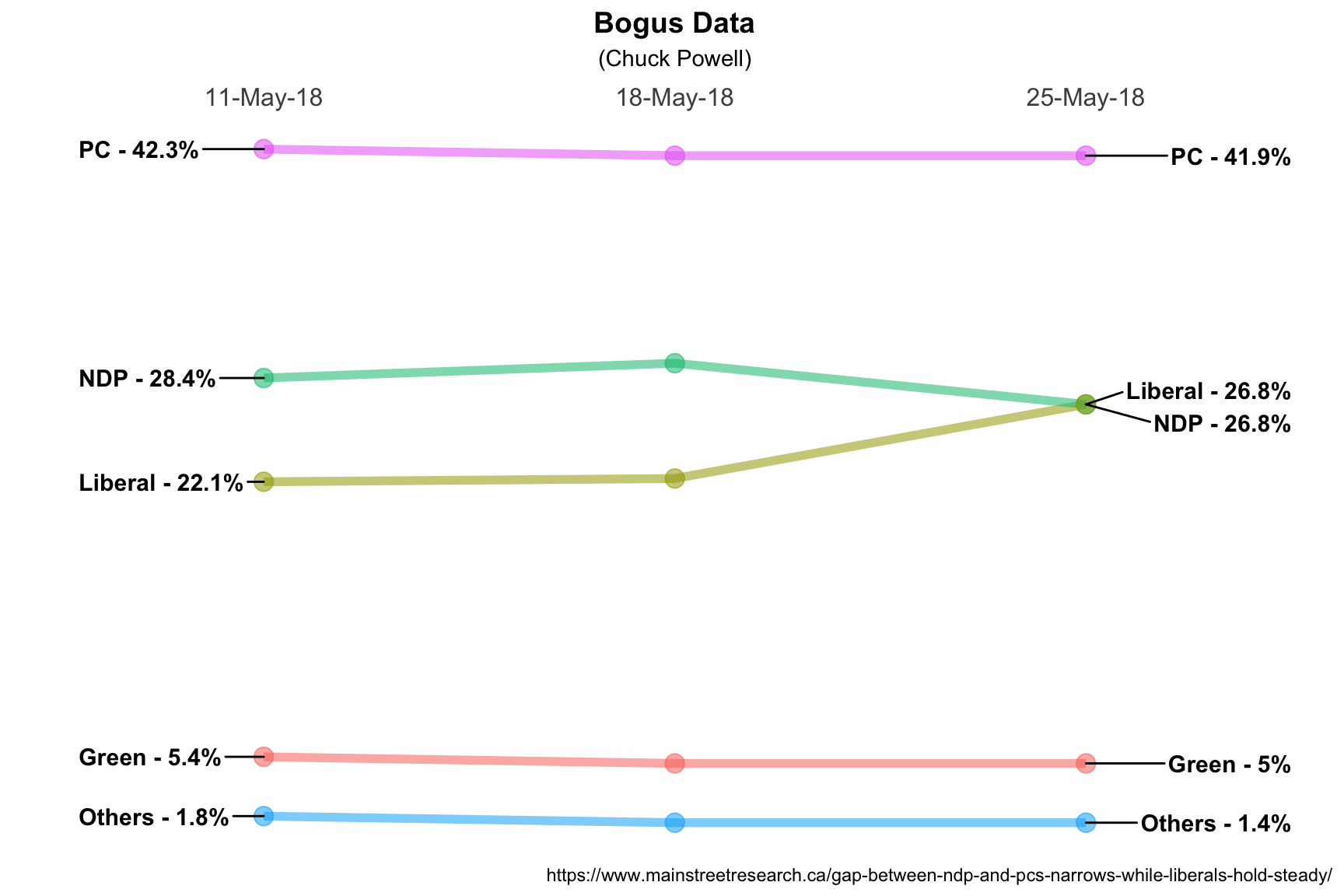
Very nice! We have confirmed that our solution works for more than two dates without any additional changes and we have found a solution to the label overlap issue. In a little while we’ll talk about labeling the data points in the center (if we want to).
Before we move on let’s make our life a little simpler. While the output plot is good it’s a lot of code to produce one graph. Let’s see if we can simplify…
Since ggplot2 objects are just regular R objects, you can put them in a list. This means you can apply all of R’s great functional programming tools. For example, if you wanted to add different geoms to the same base plot, you could put them in a list and use lapply().
But for now let’s at least take all the invariant lines of code and put them in
a list. Then when we go to plot we can just invoke the list and remain confident
we get the right formatting. For now let’s name this list something quaint and
obvious like MySpecial.
MySpecial <- list(
# move the x axis labels up top
scale_x_discrete(position = "top"),
theme_bw(),
# Format tweaks
# Remove the legend
theme(legend.position = "none"),
# Remove the panel border
theme(panel.border = element_blank()),
# Remove just about everything from the y axis
theme(axis.title.y = element_blank()),
theme(axis.text.y = element_blank()),
theme(panel.grid.major.y = element_blank()),
theme(panel.grid.minor.y = element_blank()),
# Remove a few things from the x axis and increase font size
theme(axis.title.x = element_blank()),
theme(panel.grid.major.x = element_blank()),
theme(axis.text.x.top = element_text(size=12)),
# Remove x & y tick marks
theme(axis.ticks = element_blank()),
# Format title & subtitle
theme(plot.title = element_text(size=14, face = "bold", hjust = 0.5)),
theme(plot.subtitle = element_text(hjust = 0.5))
)
summary(MySpecial)## Length Class Mode
## [1,] 17 ScaleDiscretePosition environment
## [2,] 93 theme list
## [3,] 1 theme list
## [4,] 1 theme list
## [5,] 1 theme list
## [6,] 1 theme list
## [7,] 1 theme list
## [8,] 1 theme list
## [9,] 1 theme list
## [10,] 1 theme list
## [11,] 1 theme list
## [12,] 1 theme list
## [13,] 1 theme list
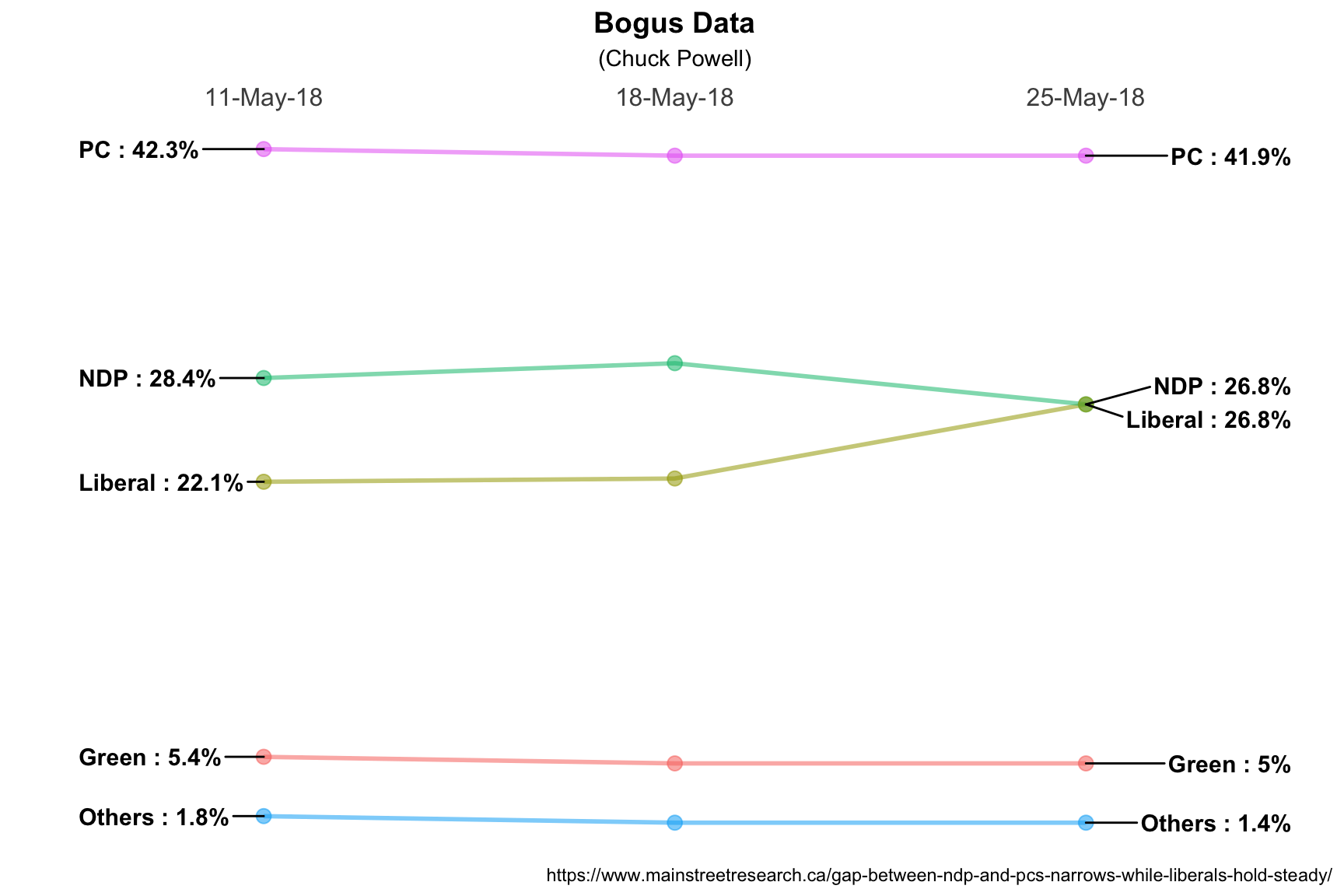
## [14,] 1 theme listMySpecial is actually an incredibly complex structure so I used the summary
function. What’s important to us is that in the future all we need to do is
include it in the ggplot command and magic happens. Perhaps another day I’ll
make it a proper function but for now I can change little things like line size
or titles and labels without worrying about the rest. So here it is with some
little things changed.
ggplot(data = moredata, aes(x = Date, y = Pct, group = Party)) +
geom_line(aes(color = Party, alpha = 1), size = 1) +
geom_point(aes(color = Party, alpha = 1), size = 3) +
geom_text_repel(data = moredata %>% filter(Date == "11-May-18"),
aes(label = paste0(Party, " : ", Pct, "%")) ,
hjust = "left",
fontface = "bold",
size = 4,
nudge_x = -.45,
direction = "y") +
geom_text_repel(data = moredata %>% filter(Date == "25-May-18"),
aes(label = paste0(Party, " : ", Pct, "%")) ,
hjust = "right",
fontface = "bold",
size = 4,
nudge_x = .5,
direction = "y") +
MySpecial +
labs(
title = "Bogus Data",
subtitle = "(Chuck Powell)",
caption = "https://www.mainstreetresearch.ca/gap-between-ndp-and-pcs-narrows-while-liberals-hold-steady/"
)
Even more complex
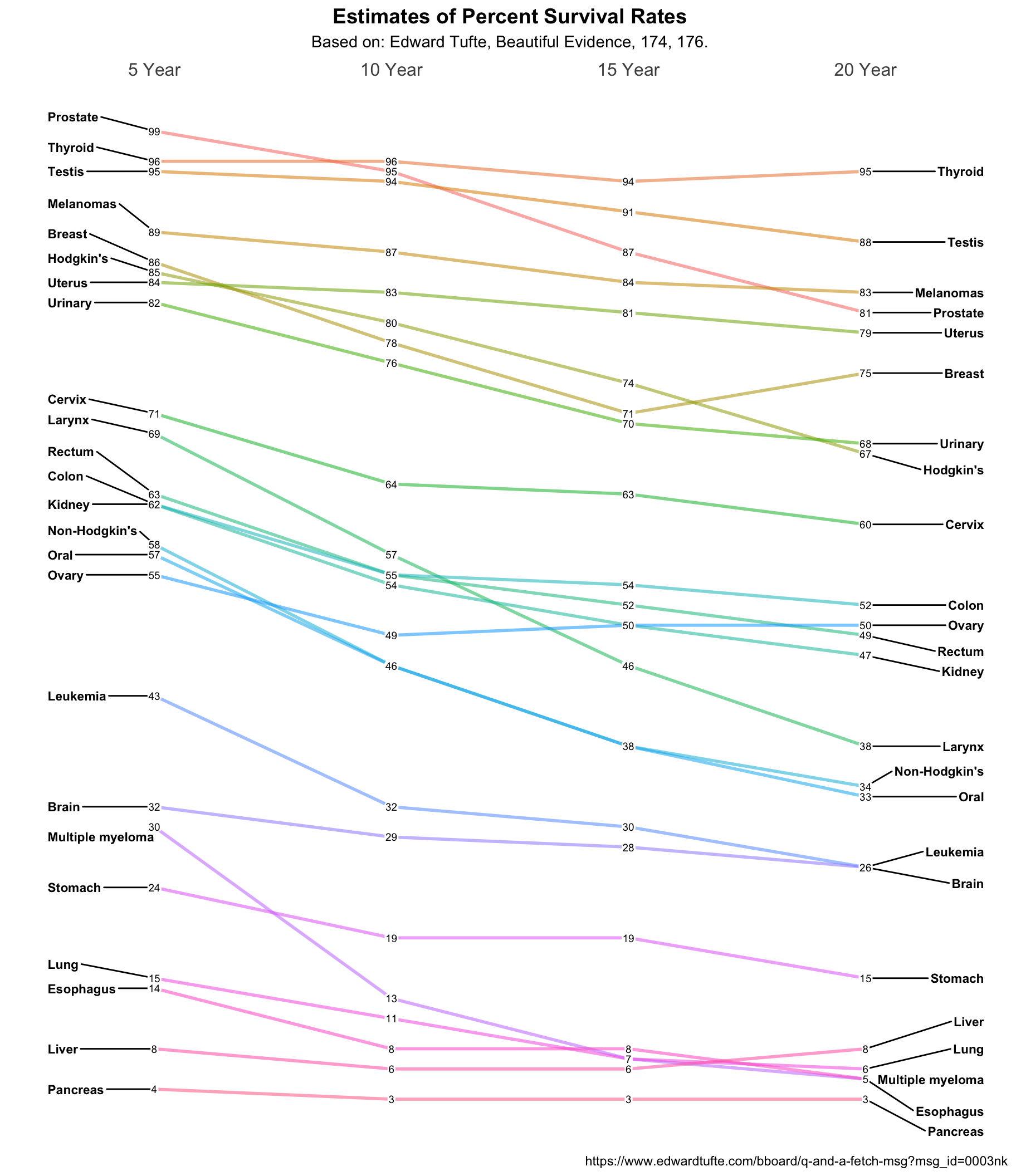
Feeling good about the solution so far I decided to press on to a much more complex problem. Thomas J. Leeper has a nice plot of Tufte’s Cancer survival slopegraph N.B. that the original Tufte is not accurate on the vertical scale. Look at Prostate and Thyroid for example since visually I would argue they should cross to reflect the data.
Let’s grab the data as laid out by Tufte.
cancer <- structure(list(Year.5 = c(99, 96, 95, 89, 86, 85, 84, 82, 71, 69, 63, 62, 62, 58, 57, 55, 43, 32, 30, 24, 15, 14, 8, 4),
Year.10 = c(95, 96, 94, 87, 78, 80, 83, 76, 64, 57, 55, 54, 55, 46, 46, 49, 32, 29, 13, 19, 11, 8, 6, 3),
Year.15 = c(87, 94, 91, 84, 71, 74, 81, 70, 63, 46, 52, 50, 54, 38, 38, 50, 30, 28, 7, 19, 7, 8, 6, 3),
Year.20 = c(81, 95, 88, 83, 75, 67, 79, 68, 60, 38, 49, 47, 52, 34, 33, 50, 26, 26, 5, 15, 6, 5, 8, 3)),
class = "data.frame",
row.names = c("Prostate", "Thyroid", "Testis", "Melanomas", "Breast", "Hodgkin's", "Uterus", "Urinary", "Cervix", "Larynx", "Rectum", "Kidney", "Colon", "Non-Hodgkin's", "Oral", "Ovary", "Leukemia", "Brain", "Multiple myeloma", "Stomach", "Lung", "Esophagus", "Liver", "Pancreas"))
str(cancer)## 'data.frame': 24 obs. of 4 variables:
## $ Year.5 : num 99 96 95 89 86 85 84 82 71 69 ...
## $ Year.10: num 95 96 94 87 78 80 83 76 64 57 ...
## $ Year.15: num 87 94 91 84 71 74 81 70 63 46 ...
## $ Year.20: num 81 95 88 83 75 67 79 68 60 38 ...kable(head(cancer,10)) %>%
kable_styling(bootstrap_options = c("striped", "hover", "condensed", "responsive"))| Year.5 | Year.10 | Year.15 | Year.20 | |
|---|---|---|---|---|
| Prostate | 99 | 95 | 87 | 81 |
| Thyroid | 96 | 96 | 94 | 95 |
| Testis | 95 | 94 | 91 | 88 |
| Melanomas | 89 | 87 | 84 | 83 |
| Breast | 86 | 78 | 71 | 75 |
| Hodgkin’s | 85 | 80 | 74 | 67 |
| Uterus | 84 | 83 | 81 | 79 |
| Urinary | 82 | 76 | 70 | 68 |
| Cervix | 71 | 64 | 63 | 60 |
| Larynx | 69 | 57 | 46 | 38 |
There, we have it in a neat data frame but not organized as we need it. Not unusual, and an opportunity to use some other tools from broom and reshape2. Let’s do the following:
- Let’s transpose the data with
t - Let’s use
broom::fix_data_frameto get valid column names and convert rownames to a proper column all in one function. Right now the types of cancer are nothing but rownames. - Use
reshape2::meltto take our transposed dataframe and convert it to long format so we can send it off toggplot. Along the way we’ll rename the resulting dataframenewcancerwith columns namedYear,TypeandSurvival.
# stepping through for demonstration purposes
t(cancer) # returns a matrix## Prostate Thyroid Testis Melanomas Breast Hodgkin's Uterus Urinary
## Year.5 99 96 95 89 86 85 84 82
## Year.10 95 96 94 87 78 80 83 76
## Year.15 87 94 91 84 71 74 81 70
## Year.20 81 95 88 83 75 67 79 68
## Cervix Larynx Rectum Kidney Colon Non-Hodgkin's Oral Ovary Leukemia
## Year.5 71 69 63 62 62 58 57 55 43
## Year.10 64 57 55 54 55 46 46 49 32
## Year.15 63 46 52 50 54 38 38 50 30
## Year.20 60 38 49 47 52 34 33 50 26
## Brain Multiple myeloma Stomach Lung Esophagus Liver Pancreas
## Year.5 32 30 24 15 14 8 4
## Year.10 29 13 19 11 8 6 3
## Year.15 28 7 19 7 8 6 3
## Year.20 26 5 15 6 5 8 3broom::fix_data_frame(
t(cancer),
newcol = "Year") # make it a dataframe with Year as a proper column## Warning: This function is deprecated as of broom 0.7.0 and will be removed from
## a future release. Please see tibble::as_tibble().## # A tibble: 4 x 25
## Year Prostate Thyroid Testis Melanomas Breast Hodgkin.s Uterus Urinary Cervix
## <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 Year… 99 96 95 89 86 85 84 82 71
## 2 Year… 95 96 94 87 78 80 83 76 64
## 3 Year… 87 94 91 84 71 74 81 70 63
## 4 Year… 81 95 88 83 75 67 79 68 60
## # … with 15 more variables: Larynx <dbl>, Rectum <dbl>, Kidney <dbl>,
## # Colon <dbl>, Non.Hodgkin.s <dbl>, Oral <dbl>, Ovary <dbl>, Leukemia <dbl>,
## # Brain <dbl>, Multiple.myeloma <dbl>, Stomach <dbl>, Lung <dbl>,
## # Esophagus <dbl>, Liver <dbl>, Pancreas <dbl>reshape2::melt(
broom::fix_data_frame(
t(cancer),
newcol = "Year"),
id="Year",
variable.name="Type",
value.name = "Survival") # melt it to long form## Warning: This function is deprecated as of broom 0.7.0 and will be removed from
## a future release. Please see tibble::as_tibble().## Year Type Survival
## 1 Year.5 Prostate 99
## 2 Year.10 Prostate 95
## 3 Year.15 Prostate 87
## 4 Year.20 Prostate 81
## 5 Year.5 Thyroid 96
## 6 Year.10 Thyroid 96
## 7 Year.15 Thyroid 94
## 8 Year.20 Thyroid 95
## 9 Year.5 Testis 95
## 10 Year.10 Testis 94
## 11 Year.15 Testis 91
## 12 Year.20 Testis 88
## 13 Year.5 Melanomas 89
## 14 Year.10 Melanomas 87
## 15 Year.15 Melanomas 84
## 16 Year.20 Melanomas 83
## 17 Year.5 Breast 86
## 18 Year.10 Breast 78
## 19 Year.15 Breast 71
## 20 Year.20 Breast 75
## 21 Year.5 Hodgkin.s 85
## 22 Year.10 Hodgkin.s 80
## 23 Year.15 Hodgkin.s 74
## 24 Year.20 Hodgkin.s 67
## 25 Year.5 Uterus 84
## 26 Year.10 Uterus 83
## 27 Year.15 Uterus 81
## 28 Year.20 Uterus 79
## 29 Year.5 Urinary 82
## 30 Year.10 Urinary 76
## 31 Year.15 Urinary 70
## 32 Year.20 Urinary 68
## 33 Year.5 Cervix 71
## 34 Year.10 Cervix 64
## 35 Year.15 Cervix 63
## 36 Year.20 Cervix 60
## 37 Year.5 Larynx 69
## 38 Year.10 Larynx 57
## 39 Year.15 Larynx 46
## 40 Year.20 Larynx 38
## 41 Year.5 Rectum 63
## 42 Year.10 Rectum 55
## 43 Year.15 Rectum 52
## 44 Year.20 Rectum 49
## 45 Year.5 Kidney 62
## 46 Year.10 Kidney 54
## 47 Year.15 Kidney 50
## 48 Year.20 Kidney 47
## 49 Year.5 Colon 62
## 50 Year.10 Colon 55
## 51 Year.15 Colon 54
## 52 Year.20 Colon 52
## 53 Year.5 Non.Hodgkin.s 58
## 54 Year.10 Non.Hodgkin.s 46
## 55 Year.15 Non.Hodgkin.s 38
## 56 Year.20 Non.Hodgkin.s 34
## 57 Year.5 Oral 57
## 58 Year.10 Oral 46
## 59 Year.15 Oral 38
## 60 Year.20 Oral 33
## 61 Year.5 Ovary 55
## 62 Year.10 Ovary 49
## 63 Year.15 Ovary 50
## 64 Year.20 Ovary 50
## 65 Year.5 Leukemia 43
## 66 Year.10 Leukemia 32
## 67 Year.15 Leukemia 30
## 68 Year.20 Leukemia 26
## 69 Year.5 Brain 32
## 70 Year.10 Brain 29
## 71 Year.15 Brain 28
## 72 Year.20 Brain 26
## 73 Year.5 Multiple.myeloma 30
## 74 Year.10 Multiple.myeloma 13
## 75 Year.15 Multiple.myeloma 7
## 76 Year.20 Multiple.myeloma 5
## 77 Year.5 Stomach 24
## 78 Year.10 Stomach 19
## 79 Year.15 Stomach 19
## 80 Year.20 Stomach 15
## 81 Year.5 Lung 15
## 82 Year.10 Lung 11
## 83 Year.15 Lung 7
## 84 Year.20 Lung 6
## 85 Year.5 Esophagus 14
## 86 Year.10 Esophagus 8
## 87 Year.15 Esophagus 8
## 88 Year.20 Esophagus 5
## 89 Year.5 Liver 8
## 90 Year.10 Liver 6
## 91 Year.15 Liver 6
## 92 Year.20 Liver 8
## 93 Year.5 Pancreas 4
## 94 Year.10 Pancreas 3
## 95 Year.15 Pancreas 3
## 96 Year.20 Pancreas 3# all those steps in one long line saved to a new dataframe
newcancer <- reshape2::melt(broom::fix_data_frame(t(cancer), newcol = "Year"), id="Year", variable.name="Type", value.name = "Survival")## Warning: This function is deprecated as of broom 0.7.0 and will be removed from
## a future release. Please see tibble::as_tibble().Now we have whipped the data into the shape we need it. 96 rows with the three
columns we want to plot, Year, Type, and Survival. If you look at the data
though, you’ll notice two small faults. First, Year is not a factor. The
plot will work but have an annoying limitation. Since “Year.5” is a character
string it will be ordered after all the other years. We could fix that on the
fly within our ggplot call but I find it cleaner and more understandable if I
take care of that first. I’ll use the factor function from base R to
accomplish that and while I’m at it make the values nicer looking. Second in
three cases R changed cancer type names because they couldn’t be column names
in a dataframe. I’ll use forcats::fct_recode to make them look better.
newcancer$Year <- factor(newcancer$Year,
levels = c("Year.5", "Year.10", "Year.15", "Year.20"),
labels = c("5 Year","10 Year","15 Year","20 Year"),
ordered = TRUE)
newcancer$Type <- forcats::fct_recode(newcancer$Type,
"Hodgkin's" = "Hodgkin.s",
"Non-Hodgkin's" = "Non.Hodgkin.s",
"Multiple myeloma" = "Multiple.myeloma")
head(newcancer)## Year Type Survival
## 1 5 Year Prostate 99
## 2 10 Year Prostate 95
## 3 15 Year Prostate 87
## 4 20 Year Prostate 81
## 5 5 Year Thyroid 96
## 6 10 Year Thyroid 96Now that we have the data the way we want it we can make our slopegraph. Some of
the necessary changes are obvious x = Year, y = Survival and group = Type
for example. Since there are a lot of plotted lines I’ve reduced the weight or
size of the individual lines. We no longer want to plot the big round points,
we’re going to substitute in the actual numbers, so that line gets commented
out. The left and right labels require no change and geom_text_repel will keep
them from overlapping which is almost inevitable given the data. To put the
actual survival numbers on the plot we’ll turn to geom_label. It’s like
geom_text only it puts a label box around the text. We’ll choose a smallish
size, minimize the amount of padding, and make the border of the box invisible.
The end result is what we want. It overlays on top of the lines we’ve already
plotted and the invisible padding gives us just enough room.
ggplot(data = newcancer, aes(x = Year, y = Survival, group = Type)) +
geom_line(aes(color = Type, alpha = 1), size = 1) +
# geom_point(aes(color = Type, alpha = .1), size = 4) +
geom_text_repel(data = newcancer %>% filter(Year == "5 Year"),
aes(label = Type) ,
hjust = "left",
fontface = "bold",
size = 3,
nudge_x = -.45,
direction = "y") +
geom_text_repel(data = newcancer %>% filter(Year == "20 Year"),
aes(label = Type) ,
hjust = "right",
fontface = "bold",
size = 3,
nudge_x = .5,
direction = "y") +
geom_label(aes(label = Survival),
size = 2.5,
label.padding = unit(0.05, "lines"),
label.size = 0.0) +
MySpecial +
labs(
title = "Estimates of Percent Survival Rates",
subtitle = "Based on: Edward Tufte, Beautiful Evidence, 174, 176.",
caption = "https://www.edwardtufte.com/bboard/q-and-a-fetch-msg?msg_id=0003nk"
)
Done for now
There was some polite interest and it was a good chance to practice my functional programming skills so I decided to see if I could make a decent R function from what I had learned. It’s in pretty good shape so I just pushed an update to CRAN. I hope you’ve found this useful. I am always open to comments, corrections and suggestions.
Chuck (ibecav at gmail dot com)
