Pairwise Bayesian Comparisons - even faster
This post builds upon two earlier posts:
- Comparing Frequentist, Bayesian and Simulation methods and conclusions
- More Bayes and multiple comparisons
Background
This all started with a nice post from Anindya Mozumdar on the R Bloggers feed. The topic material was fun for me (analyzing the performance of male 100m sprinters and the fastest man on earth), as well as exploring bayesian methods.
Last post in this series I made use of one of the nice features about a Bayesian approach - we don’t have to worry nearly as much about the multiple comparisons issue [Gelman, Hill, Yajima (2012)]. But, quite frankly, the code was very ugly with a lot of repetition and cutting and pasting. In this post I want to clean that all up. So let’s load the necessary libraries.
library(rvest) # to ha"rvest" the web page
library(tidyverse) # using readr, dplyr, and purrr
library(ggstatsplot)
library(BayesFactor)Next let’s duplicate Anindya’s earlier work and scrape the Track and Field
All-Time Performances webpage to
get the data. One change I’m making is to remove n_max = 3263 which is
unnecessary and was preventing grabbing the newer race results from summer
2019.
male_100_html <-
read_html("http://www.alltime-athletics.com/m_100ok.htm")
male_100_pres <- male_100_html %>%
html_nodes(xpath = "//pre")
male_100_htext <- male_100_pres %>%
html_text()
male_100_htext <- male_100_htext[[1]]
male_100 <- read_fwf(
male_100_htext,
skip = 1,
# n_max = 3263, # n_max removed to cpture newer races
col_types = cols(.default = col_character()),
col_positions = fwf_positions(
c(1, 16, 27, 35, 66, 74, 86, 93, 123),
c(15, 26, 34, 65, 73, 85, 92, 122, NA)
)
)
male_100 <- male_100 %>%
select(X2, X4) %>%
transmute(timing = X2, runner = X4) %>%
mutate(
timing = gsub("A", "", timing),
timing = as.numeric(timing)
)
# 3267 as of July 8, 2019
nrow(male_100) # if you're cautious you can check against the webpage## [1] 3398Let’s focus on the top 6 runners who have more than 40 race results recorded. We’ll make an effort throughout this post to capture the parameters we use and store them as variables and use the names. If you choose to replicate this post on your own you should be able to change the parameters below and see how the results vary based upon your choices (for example the top 5 or 10 runners or more than 30 races).
numbraces <- 40
howmanyrunners <- 6Having made our selections let’s use a series a dplyr commands piped %>%
together to create a character vector called orderbymean which contains the
names of the 6 runners who meet our criteria. We can use this
vector to filter our dataframe down to just the 6 we want with
a filter(runner %in% orderbymean) statement as well as force the factor levels
of runner to be in mean order with factor(male_100$runner, levels = orderbymean).
orderbymean <- male_100 %>%
group_by(runner) %>%
summarise(avgtime = mean(timing), races = n()) %>%
arrange(avgtime) %>%
filter(races >= numbraces) %>%
top_n(-howmanyrunners, avgtime) %>%
pull(runner) %>%
as.character()## `summarise()` ungrouping output (override with `.groups` argument)orderbymean## [1] "Usain Bolt" "Asafa Powell" "Tyson Gay" "Justin Gatlin" "Yohan Blake" "Maurice Greene"male_100 <- male_100 %>%
filter(runner %in% orderbymean) %>%
mutate_if(is.character, as.factor) %>%
droplevels()
male_100$runner <-
factor(
male_100$runner,
levels = orderbymean
)
glimpse(male_100)## Rows: 546
## Columns: 2
## $ timing <dbl> 9.58, 9.63, 9.69, 9.69, 9.69, 9.71, 9.72, 9.72, 9.74, 9.74, 9.75, 9.75, 9.75, 9.75, 9.76, 9.…
## $ runner <fct> Usain Bolt, Usain Bolt, Usain Bolt, Tyson Gay, Yohan Blake, Tyson Gay, Usain Bolt, Asafa Pow…levels(male_100$runner)## [1] "Usain Bolt" "Asafa Powell" "Tyson Gay" "Justin Gatlin" "Yohan Blake" "Maurice Greene"Okay we now have the 546 relevant times for the 6
runners we’re focusing on. The focus of this post is to build
the bayesian equivalent of a frequentist’s pairwise comparisons test across all
the unique runner pairings pairwise.t.test(x = male_100$timing, g = male_100$runner, p.adjust.method = "holm"). As I mentioned in an earlier
post this
would be the logical next step after conducting a oneway ANOVA of timing ~ runner or it’s bayesian equivalent BayesFactor::anovaBF(timing ~ runner, male_100)
##
## Pairwise comparisons using t tests with pooled SD
##
## data: male_100$timing and male_100$runner
##
## Usain Bolt Asafa Powell Tyson Gay Justin Gatlin Yohan Blake
## Asafa Powell 0.04034 - - - -
## Tyson Gay 0.02510 1.00000 - - -
## Justin Gatlin 0.00049 0.78167 1.00000 - -
## Yohan Blake 0.00047 0.47698 1.00000 1.00000 -
## Maurice Greene 6.9e-05 0.18338 1.00000 1.00000 1.00000
##
## P value adjustment method: holmOur Bayes equivalent to this matrix won’t report “p values” but rather the Bayes Factor associated with the pairing. Unlike the frequentist’s “reject/don’t reject” criteria, the BF we report will indicate what the data provide as odds that our hypothesis is correct. We’ll build it methodically and with an eye towards code that is easily reused in the future.
So, quick quiz, how many unique pair combinations are there for our 6
runners? The order of the pair doesn’t matter at this juncture
we’re looking for the number of possible head to head races among these 6
runners. If you’re like me you don’t know the answer to that off
of the top of your head and it can be tedious figuring it out, so let’s let the
computer always calculate it for us, and tell us. For this case it’s
15.
Then we can use combn to take the 6 names and show us what those pairings are
e.g., “Usain Bolt, Asafa Powell”. The result is a matrix of 15 columns
(all the pairings) and two rows (the names of the runners for the pairings).
Just to make it easier to see I’ve added a t() so the display is vertical
and you can see the pairs.
numberofpairings <- factorial(howmanyrunners) /
(factorial(2) * factorial(howmanyrunners - 2))
numberofpairings## [1] 15t(combn(orderbymean, 2))## [,1] [,2]
## [1,] "Usain Bolt" "Asafa Powell"
## [2,] "Usain Bolt" "Tyson Gay"
## [3,] "Usain Bolt" "Justin Gatlin"
## [4,] "Usain Bolt" "Yohan Blake"
## [5,] "Usain Bolt" "Maurice Greene"
## [6,] "Asafa Powell" "Tyson Gay"
## [7,] "Asafa Powell" "Justin Gatlin"
## [8,] "Asafa Powell" "Yohan Blake"
## [9,] "Asafa Powell" "Maurice Greene"
## [10,] "Tyson Gay" "Justin Gatlin"
## [11,] "Tyson Gay" "Yohan Blake"
## [12,] "Tyson Gay" "Maurice Greene"
## [13,] "Justin Gatlin" "Yohan Blake"
## [14,] "Justin Gatlin" "Maurice Greene"
## [15,] "Yohan Blake" "Maurice Greene"Purrring right along
Now we can grab the output from combn and create two separate vectors,
runner1 and runner2. We’ll take those vectors and create a series of purrr
statements using pipes.
The first is a
map2which takesrunner1andrunner2and creates a list of 15 dataframes, one for each pairing. The anonymousfunction(a, b)is simply an organized way of working our way through the 15 pairings. Filtering and dropping levels and explicitly converting to a dataframe becauseBayesFactor::ttestBFwill generate a warning if you pass it a tibble.Next we
purrr::mapthe list we just created (.x = .) and call thettestBFfunction. For each item in the list of 15 dataframes (one for each runner pairing) it runs withformula = timing ~ runner, the dataframe we passeddata = ., and in this case we have deliberately specified a directional hypothesisnullInterval = c(-Inf, 0)(it’s “-Inf” because the faster runner has a smaller timing). See the excellent BayesFactor documentation here for a more complete explanation of directional hypothesis testing.We started with two vectors
runner1andrunner2aftermap2we had a list of 15 dataframes. Now after the firstmappipe we have a list of 15 Bayes Factor objects (that’s whatttestBFgenerates). We’ll immediately pipe (%>%) that list (.x = .) into anothermapwhere we’ll invoke theextractBFfunction.extractBFproduces a dataframe, in this case with 2 rows, one with the rowname “Alt., r=0.707 -Inf<d<0” and the other with the name “Alt., r=0.707 !(-Inf<d<0)” which we don’t need.Now we have a list of 15 dataframes, this time containing the results of our
ttestBF. We want the row in each of them that is named “Alt., r=0.707 -Inf<d<0” and the column named “bf”, which is where the bayes factor itself is stored. So what we want is a list of 15 real numbers. So this time we’ll usemap_dblto let it know we want a list of 15 numbers,map_dbl(.x = ., ~ .["Alt., r=0.707 -Inf<d<0", "bf"]).The final pipe simply does some trivial rounding.
runner1 <- combn(orderbymean, 2)[1, ]
runner1## [1] "Usain Bolt" "Usain Bolt" "Usain Bolt" "Usain Bolt" "Usain Bolt" "Asafa Powell"
## [7] "Asafa Powell" "Asafa Powell" "Asafa Powell" "Tyson Gay" "Tyson Gay" "Tyson Gay"
## [13] "Justin Gatlin" "Justin Gatlin" "Yohan Blake"runner2 <- combn(orderbymean, 2)[2, ]
runner2## [1] "Asafa Powell" "Tyson Gay" "Justin Gatlin" "Yohan Blake" "Maurice Greene" "Tyson Gay"
## [7] "Justin Gatlin" "Yohan Blake" "Maurice Greene" "Justin Gatlin" "Yohan Blake" "Maurice Greene"
## [13] "Yohan Blake" "Maurice Greene" "Maurice Greene"bfresults <- map2(
runner1,
runner2,
function(a, b)
male_100 %>%
filter(runner %in% c(a, b)) %>%
droplevels() %>%
as.data.frame()
) %>%
map(.x = ., ~ ttestBF(
formula = timing ~ runner,
data = .,
nullInterval = c(-Inf, 0)
)) %>%
map(.x = ., ~ extractBF(x = .)) %>%
map_dbl(.x = ., ~ .["Alt., r=0.707 -Inf<d<0", "bf"]) %>%
round(., digits = 4)
bfresults## [1] 11.4819 9.4539 175.1222 211.2223 2709.0974 0.3083 1.0125 2.1714 7.6600 0.3007
## [11] 0.5028 0.8574 0.2476 0.3487 0.2241To some, the complex set of steps that leads to bfresults may look daunting.
I’d be a liar if I tried to say I wrote all those lines in one pass and got
everything right. My suggestion is that as you build the pipeline you work step
by step producing intermediate objects. Once you get the individual steps
correct it’s trivial to join them using %>% and .x = ..
Now that we have our 15 bayes factors for each of the 15 pairings of runners we
should probably join them together into one neat dataframe resultsdf that lays
everything out for us. Based on the data available we would read line #5 as the
odds are 2709:1 that Usain is faster than Maurice.
resultsdf <-
data.frame(
Runner1 = runner1,
Runner2 = runner2,
oddsfaster = bfresults
)
resultsdf## Runner1 Runner2 oddsfaster
## 1 Usain Bolt Asafa Powell 11.4819
## 2 Usain Bolt Tyson Gay 9.4539
## 3 Usain Bolt Justin Gatlin 175.1222
## 4 Usain Bolt Yohan Blake 211.2223
## 5 Usain Bolt Maurice Greene 2709.0974
## 6 Asafa Powell Tyson Gay 0.3083
## 7 Asafa Powell Justin Gatlin 1.0125
## 8 Asafa Powell Yohan Blake 2.1714
## 9 Asafa Powell Maurice Greene 7.6600
## 10 Tyson Gay Justin Gatlin 0.3007
## 11 Tyson Gay Yohan Blake 0.5028
## 12 Tyson Gay Maurice Greene 0.8574
## 13 Justin Gatlin Yohan Blake 0.2476
## 14 Justin Gatlin Maurice Greene 0.3487
## 15 Yohan Blake Maurice Greene 0.2241The Matrix reloaded (still waiting for #4)
Now that we have our resultsdf we can continue about the business of comparing
the frequentist results of paired t-tests with their bayesian counterparts.
Imagine that we have just completed a Oneway ANOVA of timing ~ runner (I’ll
show the results in a bit). Given significant results of the omnibuds F test
our next step is likely to run all the pairwise comparisons with some sort of
correction for multiple comparisons like pairwise.t.test. (see this post for
a review)
The results are almost always given as a matrix often without repeating one of
the diagonals. The results tell us that we can reject the null hypothesis that
the runners have the same time for Usain Bolt versus all the other competitors.
But it doesn’t allow us to make any statements about how different (despite
the temptation inherent in the very different p values). It supplies almost no
information about the other pairings, just that we can not reject the null.
pairwise.t.test(
x = male_100$timing,
g = male_100$runner,
p.adjust.method = "holm"
)##
## Pairwise comparisons using t tests with pooled SD
##
## data: male_100$timing and male_100$runner
##
## Usain Bolt Asafa Powell Tyson Gay Justin Gatlin Yohan Blake
## Asafa Powell 0.04034 - - - -
## Tyson Gay 0.02510 1.00000 - - -
## Justin Gatlin 0.00049 0.78167 1.00000 - -
## Yohan Blake 0.00047 0.47698 1.00000 1.00000 -
## Maurice Greene 6.9e-05 0.18338 1.00000 1.00000 1.00000
##
## P value adjustment method: holmAnother typical way of displaying the information is graphically as demonstrated
here by ggstatsplot::ggbetweenstats.
ggbetweenstats(
data = male_100,
x = runner,
y = timing,
type = "p",
var.equal = TRUE,
pairwise.comparisons = TRUE,
pairwise.display = "all",
partial = FALSE,
effsize.type = "unbiased",
sort = "ascending",
point.jitter.height = 0,
messages = FALSE
)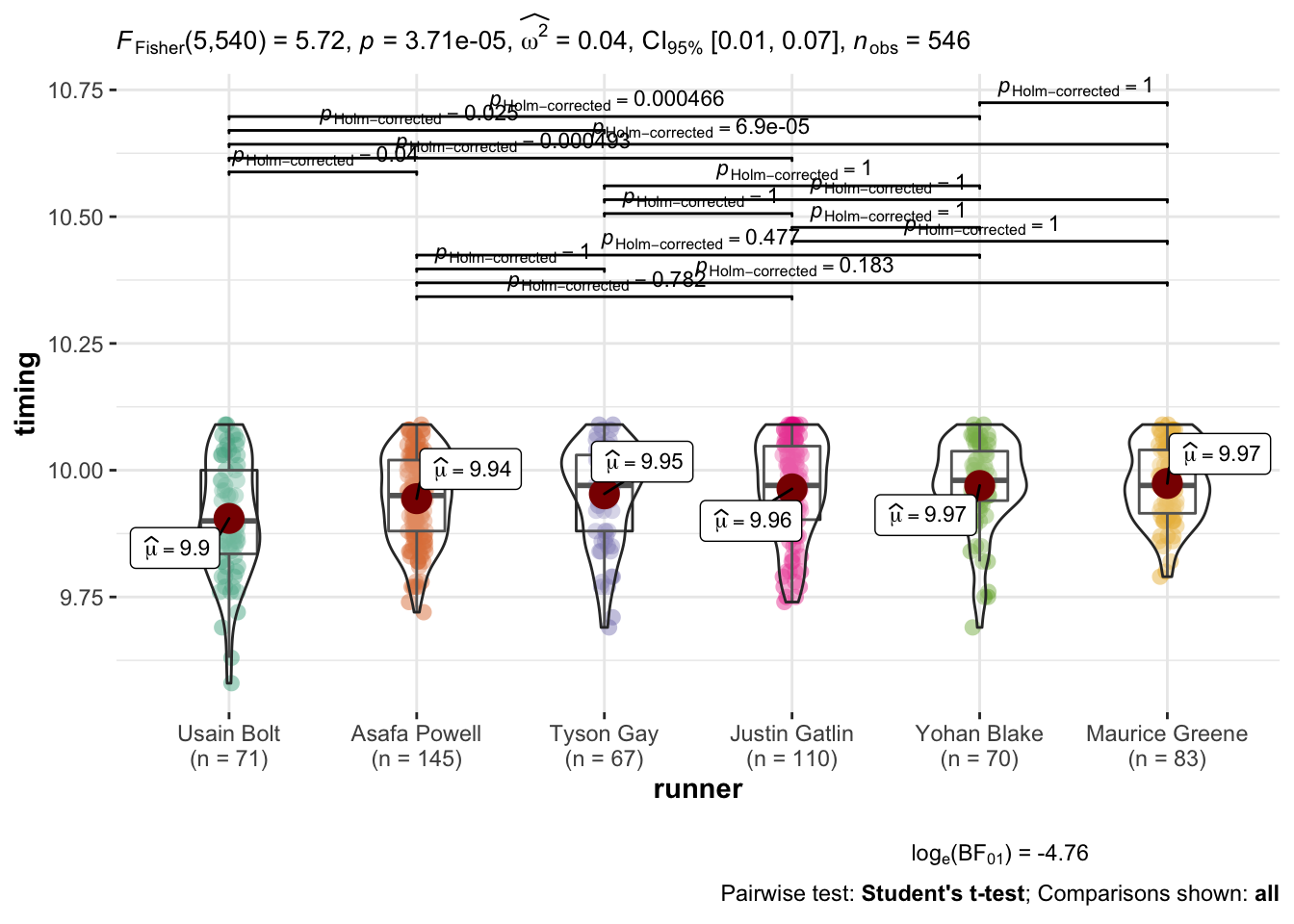
Let’s see if we can’t at least produce a similar matrix to what
pairwise.t.test yields. I’d like us to be able to do a sort of side by side
comparison of the frequentists versus bayesian results.
Step by step the process we’ll follow is:
- Use
diagto create a matrix with ones in the diagonal we’ll set the size tohowmanyrunners - Grab the runners names from
orderbymeanand populate therownamesandcolnames - Use
combnagain this time populating it with numbers (one&two) rather than the runners names - Feed those vectors into a
forloop to populate thebfmatrixwith the data fromresultsdf - To be consistent with
pairwise.t.testremove the first rowbfmatrix[-1, ]and the last columnbfmatrix[, -howmanyrunners] - Finally populate the upper triangle part of the matrix with NA
bfmatrix[upper.tri(bfmatrix)] <- NA
bfmatrix <- diag(nrow = howmanyrunners)
rownames(bfmatrix) <- orderbymean
colnames(bfmatrix) <- orderbymean
bfmatrix## Usain Bolt Asafa Powell Tyson Gay Justin Gatlin Yohan Blake Maurice Greene
## Usain Bolt 1 0 0 0 0 0
## Asafa Powell 0 1 0 0 0 0
## Tyson Gay 0 0 1 0 0 0
## Justin Gatlin 0 0 0 1 0 0
## Yohan Blake 0 0 0 0 1 0
## Maurice Greene 0 0 0 0 0 1combn(howmanyrunners, 2)## [,1] [,2] [,3] [,4] [,5] [,6] [,7] [,8] [,9] [,10] [,11] [,12] [,13] [,14] [,15]
## [1,] 1 1 1 1 1 2 2 2 2 3 3 3 4 4 5
## [2,] 2 3 4 5 6 3 4 5 6 4 5 6 5 6 6one <- combn(howmanyrunners, 2)[2, ]
two <- combn(howmanyrunners, 2)[1, ]
for (i in 1:numberofpairings) {
bfmatrix[one[i], two[i]] <- resultsdf[i, 3] # row i, column 3 which is the BF value
}
bfmatrix## Usain Bolt Asafa Powell Tyson Gay Justin Gatlin Yohan Blake Maurice Greene
## Usain Bolt 1.0000 0.0000 0.0000 0.0000 0.0000 0
## Asafa Powell 11.4819 1.0000 0.0000 0.0000 0.0000 0
## Tyson Gay 9.4539 0.3083 1.0000 0.0000 0.0000 0
## Justin Gatlin 175.1222 1.0125 0.3007 1.0000 0.0000 0
## Yohan Blake 211.2223 2.1714 0.5028 0.2476 1.0000 0
## Maurice Greene 2709.0974 7.6600 0.8574 0.3487 0.2241 1bfmatrix <- bfmatrix[-1, ]
bfmatrix <- bfmatrix[, -howmanyrunners]
bfmatrix## Usain Bolt Asafa Powell Tyson Gay Justin Gatlin Yohan Blake
## Asafa Powell 11.4819 1.0000 0.0000 0.0000 0.0000
## Tyson Gay 9.4539 0.3083 1.0000 0.0000 0.0000
## Justin Gatlin 175.1222 1.0125 0.3007 1.0000 0.0000
## Yohan Blake 211.2223 2.1714 0.5028 0.2476 1.0000
## Maurice Greene 2709.0974 7.6600 0.8574 0.3487 0.2241bfmatrix[upper.tri(bfmatrix)] <- NA
bfmatrix## Usain Bolt Asafa Powell Tyson Gay Justin Gatlin Yohan Blake
## Asafa Powell 11.4819 NA NA NA NA
## Tyson Gay 9.4539 0.3083 NA NA NA
## Justin Gatlin 175.1222 1.0125 0.3007 NA NA
## Yohan Blake 211.2223 2.1714 0.5028 0.2476 NA
## Maurice Greene 2709.0974 7.6600 0.8574 0.3487 0.2241Success! bfmatrix now looks a lot like the results of pairwise.t.test. But
we can do better. Looking at the object produced by pairwise.t.test using
str() we can see it is a list with 4 items. $method contains a simple text
string explaining what it is. $data.name is another text string that tells us
where the data came from. $p.value contains the actual p values and finally
$p.adjust.method contains which method of p adjustment (holm) we used. Since
there is no analog “adjustment method” for a bayes factor we can ignore it.
Let’s make our own list called bfpairs that mimics that structure.
str(pairwise.t.test(
x = male_100$timing,
g = male_100$runner,
p.adjust.method = "holm"
))## List of 4
## $ method : chr "t tests with pooled SD"
## $ data.name : chr "male_100$timing and male_100$runner"
## $ p.value : num [1:5, 1:5] 0.040339 0.025104 0.000493 0.000466 0.000069 ...
## ..- attr(*, "dimnames")=List of 2
## .. ..$ : chr [1:5] "Asafa Powell" "Tyson Gay" "Justin Gatlin" "Yohan Blake" ...
## .. ..$ : chr [1:5] "Usain Bolt" "Asafa Powell" "Tyson Gay" "Justin Gatlin" ...
## $ p.adjust.method: chr "holm"
## - attr(*, "class")= chr "pairwise.htest"bfpairs <- list(
method = " r = 0.707 Alt Hyp = -Inf<d<0",
data.name = "male_100$timing and male_100$runner",
p.value = bfmatrix
)
str(bfpairs)## List of 3
## $ method : chr " r = 0.707 Alt Hyp = -Inf<d<0"
## $ data.name: chr "male_100$timing and male_100$runner"
## $ p.value : num [1:5, 1:5] 11.48 9.45 175.12 211.22 2709.1 ...
## ..- attr(*, "dimnames")=List of 2
## .. ..$ : chr [1:5] "Asafa Powell" "Tyson Gay" "Justin Gatlin" "Yohan Blake" ...
## .. ..$ : chr [1:5] "Usain Bolt" "Asafa Powell" "Tyson Gay" "Justin Gatlin" ...What we’d like to do is mimic the print method for pairwise.t.test to include
little things like substituting in an em dash instead of the NAs. To do that
we need to peek at the print method with getAnywhere(print.pairwise.htest).
getAnywhere() is a life saver if you want to be able to inspect a function.
getAnywhere(print.pairwise.htest)## A single object matching 'print.pairwise.htest' was found
## It was found in the following places
## registered S3 method for print from namespace stats
## namespace:stats
## with value
##
## function (x, digits = max(1L, getOption("digits") - 5L), ...)
## {
## cat("\n\tPairwise comparisons using", x$method, "\n\n")
## cat("data: ", x$data.name, "\n\n")
## pp <- format.pval(x$p.value, digits = digits, na.form = "-")
## attributes(pp) <- attributes(x$p.value)
## print(pp, quote = FALSE, ...)
## cat("\nP value adjustment method:", x$p.adjust.method, "\n")
## invisible(x)
## }
## <bytecode: 0x7fa368192710>
## <environment: namespace:stats>Hmmmmm. Okay so we pass it a list object (the bfpairs we just created) and it
takes the sub components and puts them into the right places on the screen.
Spoiler alert, I won’t go into all the details but suffice it to say that
format.pval() is problematic for us.
It does a very nice job working with p values but p values have a different set
of characteristics than bayes factors.
Rather than modify format.pval I simply decided to use the generic format
function instead. That way the end user can specify all sorts of parameters like
the number of digits, the symbol to replace NA, and the justification etc..
Here’s what I came up with after a little bit of work. Hopefully you’ll agree it
does a reasonably good job of replicating the functionality of
print.pairwise.htest?
print.pairwise.bftest <- function(x,
digits = 2,
nsmall = 0,
width = 9,
justify = "right",
scientific = FALSE,
nareplace = "-") {
cat("\nPairwise comparisons of bayes factors with", x$method, "\n\n")
cat("data: ", x$data.name, "\n\n")
pp <- format(x$p.value,
digits = digits,
nsmall = nsmall,
width = width,
justify = justify,
scientific = scientific
)
pp <- gsub("NA", nareplace, pp)
print(pp, quote = FALSE)
cat("\n\nAnalyzed using BayesFactor::ttestBF\n")
invisible(x)
}
print.pairwise.bftest(bfpairs, digits = 1)##
## Pairwise comparisons of bayes factors with r = 0.707 Alt Hyp = -Inf<d<0
##
## data: male_100$timing and male_100$runner
##
## Usain Bolt Asafa Powell Tyson Gay Justin Gatlin Yohan Blake
## Asafa Powell 11.5 - - - -
## Tyson Gay 9.5 0.3 - - -
## Justin Gatlin 175.1 1.0 0.3 - -
## Yohan Blake 211.2 2.2 0.5 0.2 -
## Maurice Greene 2709.1 7.7 0.9 0.3 0.2
##
##
## Analyzed using BayesFactor::ttestBFNotice that bayes factors aren’t shockingly dissimilar than the conclusions you would draw from a frequentist’s perspective. I still think they are a better choice because you can talk about odds and probabilities cleanly without falling into the frequentist “traps” surrounding what rejection of the null hypothesis is. With our “new” perspective we are safe in making statements that out data strongly support some of the pairwise differences (odds of 2709 to 1 are pretty convincing) and in other cases we can now quantify that odds are it’s “anyone’s race.”
Play it again Sam
As I wrote this post I wanted to ensure that I could run the analysis on a different set of runners with minimal effort. What follows is the code minus all of the intermediate printing and explanation. The difference is this time we’ll look at the top 7 fastest sprinters and widen our analysis to anyone with at least 20 races.
male_100_html <-
read_html("http://www.alltime-athletics.com/m_100ok.htm")
male_100_pres <- male_100_html %>%
html_nodes(xpath = "//pre")
male_100_htext <- male_100_pres %>%
html_text()
male_100_htext <- male_100_htext[[1]]
male_100 <- read_fwf(
male_100_htext,
skip = 1,
col_types = cols(.default = col_character()),
col_positions = fwf_positions(
c(1, 16, 27, 35, 66, 74, 86, 93, 123),
c(15, 26, 34, 65, 73, 85, 92, 122, NA)
)
)
male_100 <- male_100 %>%
select(X2, X4) %>%
transmute(timing = X2, runner = X4) %>%
mutate(
timing = gsub("A", "", timing),
timing = as.numeric(timing)
)
numbraces <- 20
howmanyrunners <- 7
orderbymean <- male_100 %>%
group_by(runner) %>%
summarise(avgtime = mean(timing), races = n()) %>%
arrange(avgtime) %>%
filter(races >= numbraces) %>%
top_n(-howmanyrunners, avgtime) %>%
pull(runner) %>%
as.character()
male_100 <- male_100 %>%
filter(runner %in% orderbymean) %>%
mutate_if(is.character, as.factor) %>%
droplevels()
male_100$runner <-
factor(
male_100$runner,
levels = orderbymean
)
numberofpairings <- factorial(howmanyrunners) /
(factorial(2) * factorial(howmanyrunners - 2))
runner1 <- combn(orderbymean, 2)[1, ]
runner2 <- combn(orderbymean, 2)[2, ]
bfresults <- map2(
runner1,
runner2,
function(a, b)
male_100 %>%
filter(runner %in% c(a, b)) %>%
droplevels() %>%
as.data.frame()
) %>%
map(.x = ., ~ ttestBF(
formula = timing ~ runner,
data = .,
nullInterval = c(-Inf, 0)
)) %>%
map(.x = ., ~ extractBF(x = .)) %>%
map_dbl(.x = ., ~ .["Alt., r=0.707 -Inf<d<0", "bf"]) %>%
round(., digits = 4)
resultsdf <-
data.frame(
Runner1 = runner1,
Runner2 = runner2,
oddsfaster = bfresults
)
bfmatrix <- diag(nrow = howmanyrunners)
rownames(bfmatrix) <- orderbymean
colnames(bfmatrix) <- orderbymean
one <- combn(howmanyrunners, 2)[2, ]
two <- combn(howmanyrunners, 2)[1, ]
for (i in 1:numberofpairings) {
bfmatrix[one[i], two[i]] <- resultsdf[i, 3]
}
bfmatrix <- bfmatrix[-1, ]
bfmatrix <- bfmatrix[, -howmanyrunners]
bfmatrix[upper.tri(bfmatrix)] <- NA
bfpairs <- list(
method = " r = 0.707 Alt Hyp = -Inf<d<0",
data.name = "male_100$timing and male_100$runner",
p.value = bfmatrix
)
pairwise.t.test(
x = male_100$timing,
g = male_100$runner,
p.adjust.method = "holm"
)##
## Pairwise comparisons using t tests with pooled SD
##
## data: male_100$timing and male_100$runner
##
## Usain Bolt Asafa Powell Christian Coleman Tyson Gay Justin Gatlin Yohan Blake
## Asafa Powell 0.05996 - - - - -
## Christian Coleman 0.40318 1.00000 - - - -
## Tyson Gay 0.03605 1.00000 1.00000 - - -
## Justin Gatlin 0.00067 1.00000 1.00000 1.00000 - -
## Yohan Blake 0.00061 0.72843 1.00000 1.00000 1.00000 -
## Maurice Greene 8.8e-05 0.28578 1.00000 1.00000 1.00000 1.00000
##
## P value adjustment method: holmprint.pairwise.bftest(bfpairs,
digits = 3,
scientific = TRUE,
nareplace = ".")##
## Pairwise comparisons of bayes factors with r = 0.707 Alt Hyp = -Inf<d<0
##
## data: male_100$timing and male_100$runner
##
## Usain Bolt Asafa Powell Christian Coleman Tyson Gay Justin Gatlin Yohan Blake
## Asafa Powell 1.15e+01 . . . . .
## Christian Coleman 2.18e+00 2.91e-01 . . . .
## Tyson Gay 9.45e+00 3.08e-01 2.61e-01 . . .
## Justin Gatlin 1.75e+02 1.01e+00 3.89e-01 3.01e-01 . .
## Yohan Blake 2.11e+02 2.17e+00 5.93e-01 5.03e-01 2.48e-01 .
## Maurice Greene 2.71e+03 7.66e+00 9.58e-01 8.57e-01 3.49e-01 2.24e-01
##
##
## Analyzed using BayesFactor::ttestBFAnd voila! Based on our new criteria of the fastest 7 runners with at least 20 races Christian Coleman has been added to the matrix. His mean timings place him square in the middle of the pack between Tyson Gay and Justin Gatlin. But notice the BF comparing him to Usain Bolt is only about 9.5 which is smaller than Tyson Gay 2.2 and Justin Gatlin 175.1 or any of the other runners. This is likely because the BF always adjusts based upon the amount of evidence available and we only have 28 races of data available for Christian.
Remember that one of the nice features of bayesian methodology is that we can quantify support for both the hypothesis we have as well as it’s converse (what a frequentist would call the null hypothesis). So our hypothesis is that Justin Gatlin is faster than Yohan Blake but the bayes factor 2.03e-01 (.203) says that the evidence from the data is that the odds are 1 / 2.03e-01 or about 5:1 that Justin is NOT faster than Yohan. That’s a statement that can not be made when using frequentist methods.
Done
I’ve really enjoyed this series of posts. I am always open to comments, corrections and suggestions. Feel free to leave a comment in disqus or send me an email.
Chuck
This work is licensed under a Creative Commons Attribution-ShareAlike 4.0 International License

